Introduction
Input Data Format
Uploading and Processing
Obtaining an account
Upload Form
Data uploading
Processing Datasets
Project Management
Browsing Pathways
Promoter analysis
Analyzing Dataset
Changed Pathways
GO Enrichment
Functional classification
Searching
Adding New Platforms
Update Pathways and Genes
Contacts
|
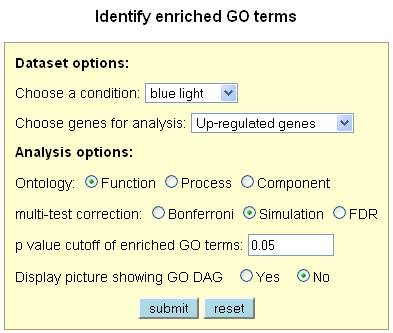
Identify Enriched GO Terms
This tool allows the user to identify over-represented (enriched) GO terms from a list of up-
and/or down-regulated genes under a specific condition. The tool was implemented based on the
GO::TermFinder perl module
(
Boyle et al., 2004) which uses the hypergeometric distribution to calculate the significance
of GO term enrichment. The tool provides three types of multiple test correction methods that
come with the GO::TermFinder module: FDR, Simulation, and Bonferroni. Significantly enriched
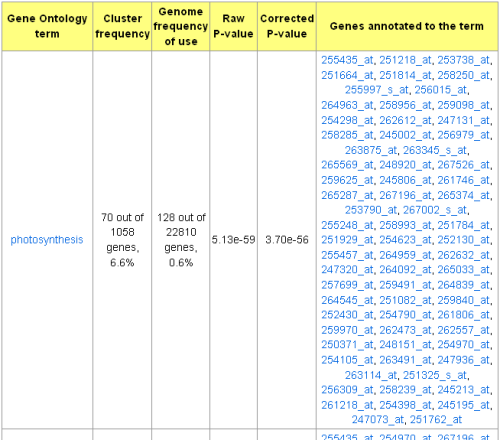
GO terms, genes annotated to each corresponding GO term, and the GO DAG picture (optional) are
included in the output page.
Note: This program may take several minutes if the number of input
genes is large.
|







