Introduction
Input Data Format
Uploading and Processing
Obtaining an account
Upload Form
Data uploading
Processing Datasets
Project Management
Browsing Pathways
Promoter analysis
Analyzing Dataset
Changed Pathways
GO Enrichment
Functional classification
Searching
Adding New Platforms
Update Pathways and Genes
Contacts
|
Promoter analysis
It has been reported that a subset of genes in the same pathway are regulated by common
transcription factors. Plant MetGenMAP provides a tool to identify over-represented motifs
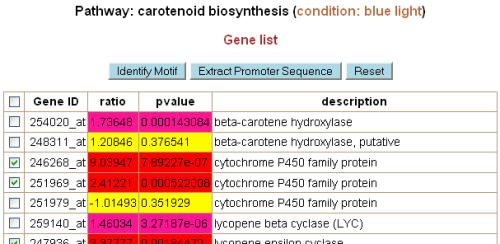
from the promoters of co-expressed genes in the same pathway. In the "Gene list" page of
each pathway, the user can extract the promoter sequences of a set of genes in the pathway
(default: genes significantly up- or down-regulated) for his/her own analysis. The user
can also identify motifs using the tool provided by the system (see below).
Motif identification
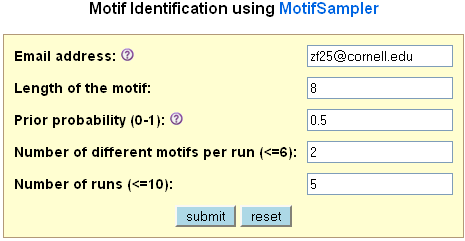
Plant MetGenMAP uses MotifSampler to identify motifs from the promoter sequences of co-expressed
genes in a particular pathway. The user can specify parameters using the input form the
system provides. For a detail explanation of these parameters, please check MotifSampler
homepage.
After motifs are identified by MotifSampler, each motif is screened against all promoter
sequences of the organism and against the promoters of the list of co-expressed genes,
respectively, using PatMatch, to identify the occurrences of the motif. Then the post-hoc
test is performed to calculate p value of the motif enrichment based on the hypergeometric
distribution.
After submitting the job, the job will run in background. Once the job is finished (it
can take several minutes), a link to the output page will be sent to the email address
the user provided (default is the email the user provided during the registration).
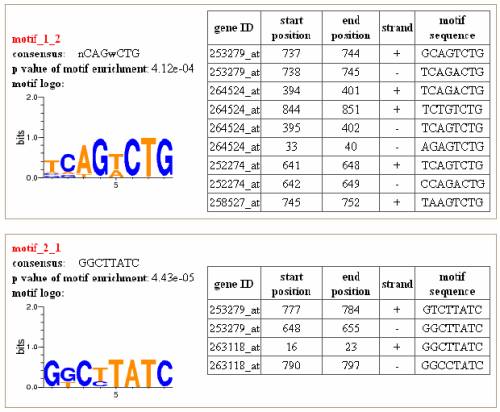
Motifs and their sequence logos, p value of the motif enrichment, as well as the positions
of the motif in the promoter of each gene, are included in the output page.
Note: Currently this tool is only available for datasets
generated from Arabidopsis and rice.
|







