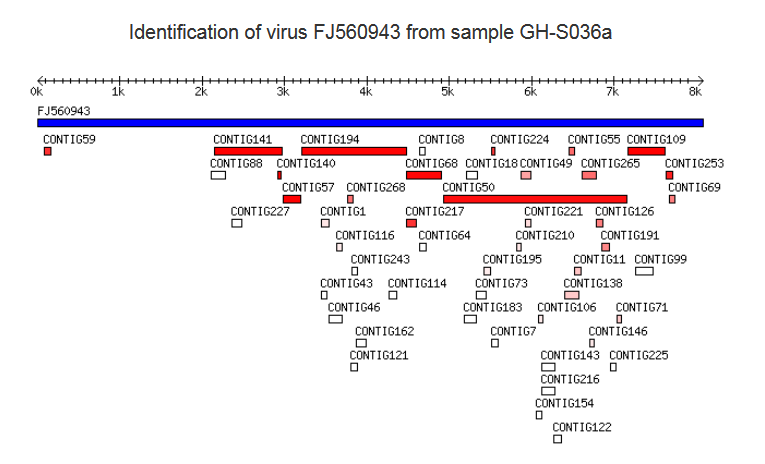
This table provides the list of reference sequences from GenBank database to which significant similarity was found in the contigs of the sample. There are several aspects to keep in mind when interpreting these data:
-
The GenBank database may contain errors and/or outdated nomenclature and therefor results should be verified by checking the reference sequences.
-
The table provides the minimum, maximum and average sequence identity to the reference sequence and this should be taken into account when interpreting the data. Significant similarity can be achieved with less than 70% sequence identity in which case it may represent a different virus than that listed. Sequence identity criteria for virus species differ between genera (more information http://www.ictvonline.org/index.asp), but generally sequence identity >90% can be considered the same species. The alignment should be further scrutinized if any of the values is below 90%. More information about evaluating alignments
-
Results will change and become more accurate over time as sequence data of newly identified viruses is added to the GenBank database and we will re-run the analysis with the same data. The version of the GenBank virus sequence database used for the current results is v206.0. (http://www.ncbi.nlm.nih.gov/news/02-17-2015-genbank-206/)